SOPHIA ANTIPOLIS, France – March 16, 2023 │ Q4 2022 Therapeutic mRNA Patent Monitor (covering mRNA design, mRNA delivery, manufacturing, storage, mRNA-based vaccines, and mRNA-based therapeutics) quarterly report is out! Let’s focus on one interesting aspect of delivery innovation.
KnowMade has developed expertise in therapeutics mRNA that allows a picture from three angles of this disruptive technology, thanks to landscape reports on mRNA vaccine, mRNA cancer therapies, and recently self-amplifying RNA vaccine. In this fast-evolving context, it is crucial to monitor the patenting activity related to therapeutic mRNA to be aware of weak signals that might be the future of this technology and understand the intellectual property position and strategy of the different players. Targeted delivery of therapeutic mRNA is one of the current challenges. Multiple studies relate to the impact of carrier material properties and routes of administration as significant parameters influencing the expression profile of mRNA therapeutics. However, in addition to these parameters, the mRNA cargo could also be modified to be expressed in specific cells. Last quarter, the patent monitoring activity (see here) showed that Moderna and CureVac are developing the use, in different ways, of endogenous miRNA silencing to target specific cells.
Targeted delivery is a key to mRNA effectiveness
For many therapeutic proteins, the realization of medical benefit is intrinsically dependent on their expression at the correct location within the body. Therefore, it is critical that mRNA is delivered into specific organs or cells to satisfy the therapy.
Surface tissues (e.g., muscle and eyes) are readily reached by local administration. For deeper organs in the body, which are hardly achieved by local delivery, systemic administration is more favorable. However, lipid-based carriers such as LNPs tend to accumulate in the liver after intravenous injection. The challenge at that point then becomes the targeted accumulation of the therapeutic mRNA at the site and cells of interest. Despite the most successful materials used for mediating delivery of mRNA being LNPs, other delivery systems are being developed, such as polymeric materials containing carriers, peptide-based nanoparticles, and even inorganic nanoparticles. Independently of the type of carrier, three kinds of targeting can be distinguished:
- Passive targeting: This approach relies on the intrinsic material properties of the delivery system, such as size, shape, stiffness, and surface charge, to target the organ and cell type of interest. Passive targeting approaches do not involve the use of an additional targeting moiety; however, they are more difficult to understand and control without a deep understanding of the tissue or cellular localization mechanisms.
- Active targeting: A ligand is used to facilitate specific binding to receptors or other cellular features highly expressed in cells of a target organ of interest. These approaches generally rely on an additional small molecule, peptide, or biologic conjugated to the delivery system, which often increases the complexity of manufacture (g., monoclonal antibodies).
- Endogenous targeting: The nanoparticle composition is engineered to bind to a distinct subset of plasma proteins upon injection, which directs it to a target organ and promotes uptake by specific cells.
Recent reviews provide summaries of existing materials’ properties and routes of administration that can lead to the desired targeted expression of an mRNA therapeutic (see fig.1)( Dilliard et al., 2023; Meyer et al., 2022).
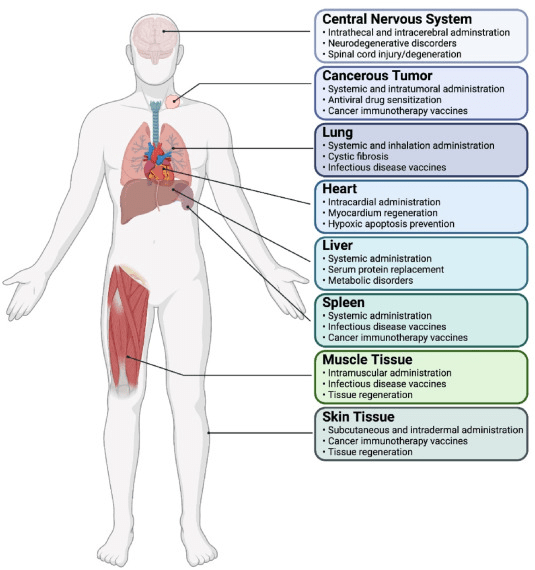
Fig.1: Schematic of the various organs and organ systems targeted for mRNA delivery
Each organ presents a summary of strategies used to target and applications realized with targeted expression of mRNA. From Meyer et al., 2022.
Using Cargo modifications for specific miRNA silencing process
With opposite strategies, CureVac and Moderna modified the mRNA sequence itself to control its expression through miRNA regulation in two patent families published in Q4 of 2022 (WO2022/233880 and WO2022/266083, respectively). Briefly, CureVac’s strategy is to turn off the expression of the therapeutic RNA in undesired cells and locations, while Moderna’s is to turn on the expression of the therapeutic mRNA in desired cells. Both strategies utilize the miRNA regulation pathway.
miRNA
MicroRNA (miRNA) are small, single-stranded, non-coding RNA molecules containing 21 to 23 nucleotides, involved in RNA silencing and post-transcriptional regulation of gene expression. miRNAs bind specifically to a complementary sequence in mRNA molecules (miRNA target sites, miRts) through base pairing. This association leads to gene silencing of the mRNA molecule either through degradation or by preventing translation. miRNA expressions patterns are highly specific in relation to external stimuli, developmental stages, and tissues. Exploiting miRNA target sites allows for the construction of targeted gene delivery platforms with various therapeutic applications (see Dhungel et al., 2018 for an example). This strategy results in repressed translation of the transgene in miRNA-positive cells, but not in miRNA-negative cells (see figure 2).
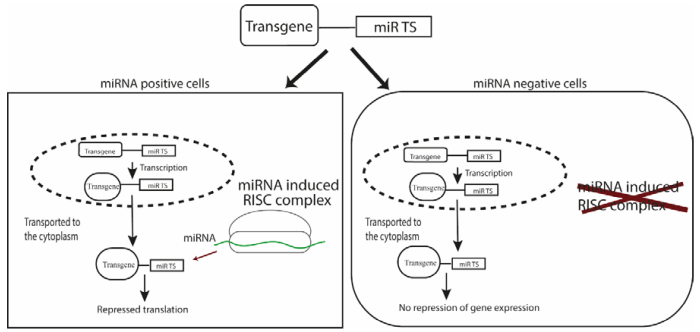
Fig. 2: Principle of miRNA mediated regulation of transgene
By introducing a binding site (TS) for a miRNA that is expressed in the target cell/tissue, a miRNA-regulated gene delivery platform for negative targeting has been built. The miRNA expressed in the target cell/tissue will inhibit transgene expression post-transcriptionally, while transgene expression in other cells remains unaffected (Dhungel et al., 2018).
CureVac strategy: Incorporation of miRts in an mRNA construct to turn off the mRNA expression in specific cells
CureVac’s strategy, as described in the recent patent application WO2022/233880, is based on the finding that the incorporation of specific miRNA target sites (miRts) before the 5′ UTR or the coding sequence of an mRNA construct can lead to a sufficient reduction of the expression from the mRNA in hepatocytes. Selected miRNA target sites are the binding site of miRNA-122, which is highly expressed in the liver, miRNA-148a, which is expressed in immune cells, the liver, and various other tissues (e.g., cerebral, heart, thymus, pancreas, renal, placenta, uterus, testis, and the hematopoietic system), miRNA-101, which is expressed in endothelial cells, epithelial cells, adipocytes, and the liver, and miRNA-192, which is expressed in the kidney, liver, and in liver disease or cancer. Experimental results show the efficiency of such miRts upstream 5’UTR or downstream 3′ UTR (see Fig. 3A). For each miRts, the repression of luciferase mRNA expression in primary human hepatocytes is more efficient when the target site is upstream of the 5’UTR, and the most efficient miRts is the one corresponding to miRNA-122. In addition, in vivo experiments confirm the organ specificity of miRNA-122, with repression of reporter mRNA in the liver and not in the spleen (see Fig. 3B).
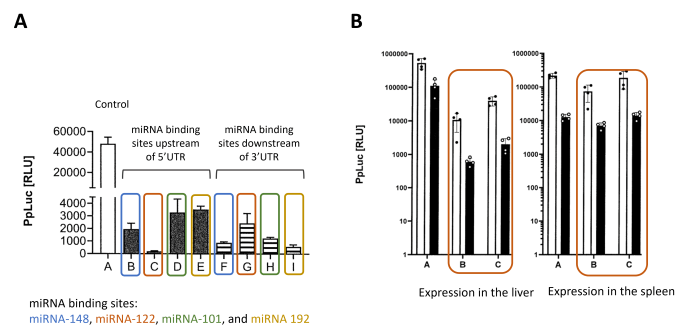
Fig. 3: Luciferase expression depending on miRNA recognition site
A. In Vitro experiment: The degree of silencing of different miRNA target sites was determined in primary human hepatocytes in an in vitro experiment. The target sites were cloned upstream of the 5’UTR or downstream of the 3’UTR.
B. In vivo Experiment: The Luciferase expression of formulated mRNA constructs containing a single miRNA target site from miRNA-122, placed upstream of the 5’UTR (B) or downstream of the 3’UTR (C) was examined in the liver and spleen of mice.
Adapted from WO2022/233880, Figures 1A, 10A and 10B detailed in examples 2 and 6.
Moderna’s tissue-specific mRNA expression strategy: Turning on mRNA expression in specific cells and locations
The strategy adopted by Moderna and described in the recent patent application WO2022/266083 is not to repress mRNA expression in off-target sites (e.g., liver), but to turn on mRNA expression in specific cells and/or locations. To achieve this, two mRNAs are co-transfected: the first is a therapeutic mRNA that needs to be expressed in a specific location, and the second is a repressor of the first mRNA expression (e.g., an mRNA encoding RNase L7Ae). As shown in Fig. 4A, the sequence of the therapeutic mRNA (encoding luciferase, Luc) contains a repressor binding site allowing for luciferase repression when the repressor is expressed. Additionally, the mRNA sequence encoding the repressor contains specific miRNA target sites (miRts) in its 3’UTR. Therefore, in organs where specific miRNAs are absent, the repressor is expressed and turns off the expression of the therapeutic mRNA. Conversely, in organs (or locations) where the therapeutic mRNA expression is desired and where specific miRNAs are present, the repressor expression is turned off, resulting in the expression of the therapeutic mRNA. Tissue-specific miRNAs include miR-122 in the liver; miR-133 or miR-206 or miR-208 in muscle; miR-17-92 or miR-126 in endothelial cells; miR-142-3p or miR-142-5p or miR-16 or miR-21 or miR-223 or miR-24 or miR-27 in myeloid cells; let-7 or miR-30c in adipose tissue; miR-1d or miR-149 in heart; miR-192 or miR-194 or miR-204 in kidney; miR142 in spleen; miR150 in lymphoid cells; and let-7 or miR-133 or miR-126 in lung epithelial cells.
This mechanism of regulation is detailed in the experimental part of the patent application which shows that, in vivo, specific expression of the reporter mRNA (luciferase) in the spleen is similar to that of the control, and a repression of such expression occurs when non-spleen-specific miRts (no miRts or 122 miRts) are added to the repressor mRNA construction.
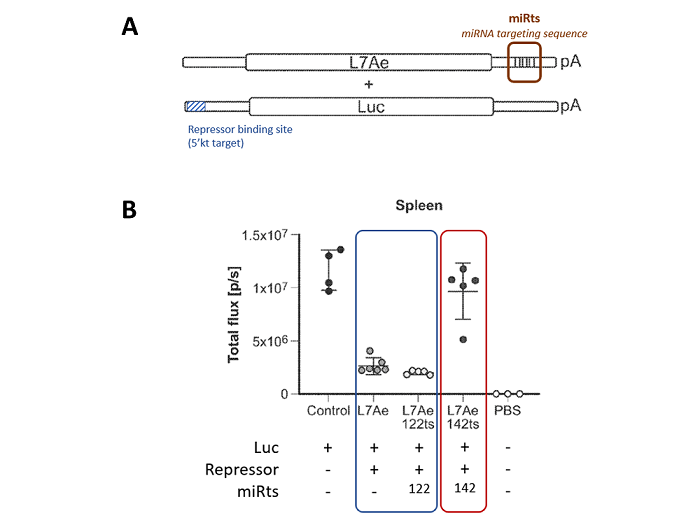
Fig. 4: in vivo Luciferase expression depending on miRNA recognition site in repressor mRNA construction
A. This diagram illustrates the two RNA construct systems used for the study. The repressor (L7Ae) is located in the 3′ untranslated region (3’UTR) of the microRNA targeting sequence, while the reporter (Luc, luciferase) is located in the 5’UTR of the repressor binding site. pA, poly A tail.
B. In vivo Experiment. BALB/c mice were injected intravenously with 0.05 mg/kg the Luc target RNA (5’ kt target) and L7Ae repressor RNA constructs (L7Ae repressor RNA with or without miRNA target sites) depicted in Fig.4A. The L7Ae repressor RNA was in molar excess (10X) of the Luc target RNA construct. The spleen of the mice was imaged by whole body luminescence 6 h post IV injection. The graph shows the level of luminescence in the spleen of mice injected with the Luc target RNA construct with Control EPO RNA or L7Ae repressor RNA or L7Ae repressor-miR122ts RNA or L7Ae repressor-miR142ts RNA. Luminescence observed for mice injected with phosphate- buffered saline (PBS) is also shown.
Adapted from WO2022/266083, Figures 6A and 6B detailed in example 4.
mRNA modifications, the future of mRNA targeted therapies?
The production of an efficient therapeutic mRNA requires the optimization of both mRNA sequence and carrier for stability, immunogenicity, and targeted delivery reasons. For now, most of the strategies developed for targeted delivery relate to carrier optimization. This necessitates a carrier for each application, and corresponding carrier manufacturing process. On the other hand, the modification of the RNA sequence by adding miRNA targeting sequences for cell/location-specific expression seems more scalable. Indeed, the development of therapeutic mRNA synthesis platforms does not depend on the sequence of the therapeutic mRNA, and, for this reason, miRts-specific sequences can be added easily. CureVac and Moderna’s approaches outlined in their recent patent applications appear promising for fine-tuned therapeutic mRNA.
Press contact
contact@knowmade.fr
Le Drakkar, 2405 route des Dolines, 06560 Valbonne Sophia Antipolis, France
www.knowmade.com
About the author
Elodie Bovier, PhD., works at KnowMade as a Patent Analyst in the field of Biotechnology and Life Sciences. She holds a PhD in genetic and molecular biology from the Paris Sud University. She also holds the Industrial Property International Studies Diploma (in Patent and Trademark & Design Law) from the CEIPI (Strasbourg, France).
About KnowMade
KnowMade is a technology intelligence and IP strategy consulting company specialized in analyzing patents and scientific publications. The company helps innovative companies, investors, and R&D organizations to understand competitive landscape, follow technological evolutions, reduce uncertainties, and identify opportunities and risks in terms of technology and intellectual property.
KnowMade’s analysts combine their strong technology expertise and in-depth knowledge of patents with powerful analytics tools and methodologies to turn patent information and scientific literature into actionable insights, providing high added value reports for decision makers working in R&D, innovation strategy, intellectual property, and marketing. Our experts provide prior art search, patent landscape analysis, freedom-to-operate analysis, IP due diligence, and monitoring services.
KnowMade has a solid expertise in Compound Semiconductors, Power Electronics, Batteries, RF Technologies & Wireless Communications, Solid-State Lighting & Display, Photonics, Memories, MEMS & Sensors, Semiconductor Packaging, Medical Devices, Medical Imaging, Microfluidics, Biotechnology, Pharmaceutics, and Agri-Food.
