SOPHIA ANTIPOLIS, France – June 16, 2022 | CureVac N.V., a German biopharmaceutical company developing medicines based on messenger ribonucleic acid (mRNA), has announced on June 08th 2022 the acquisition of Frame Cancer Therapeutics (formerly Frame Pharmaceuticals), a startup company based in Netherlands focused on advanced genomics and bioinformatics to identify both unique and shared neoantigens across different cancer types. The total consideration for the acquisition of Frame Therapeutics is valued at €32 million. It will be paid in CureVac shares. In the context of this acquisition and to understand why CureVac has acquired Frame Cancer Therapeutics, it is interesting to analyze the patent portfolio of this Dutch startup. Knowmade aims to produce such patent analyses – you may find it interesting to read our work on analyzing patent activity in healthcare.
IP analysis shows that Frame Therapeutics owns 7 patent families, filed between 2018 and 2020. Among these patent families, 4 are focused on cancer vaccines for specific cancers, i.e., breast (WO2020022899), kidney (WO2020022900), colorectal (WO2020022902) and uterine (WO2020022901) cancers. Patent family WO2020022903 is broader and concerns cancer cells.
Neoantigen-based cancer vaccines
All these patent families find their source in a piece of work carried out in 2018, also published in Scientific Reports (available here). In this work, inventors analyzed 10,186 cancer genomes from 33 tumor types and focused on the 143,444 frame-shift mutations represented in this cohort. Translation of these mutations after re-annotation to a RefSeq annotation, starting in the protein reading frame, can lead to 70,439 unique peptides that are 10 or more amino acids in length.
According to these inventions, all these vaccines are also useful for ‘off-the-shelf’ use. Indeed, inventors report that frame-shift mutations, which are mostly unique among patients and tumors, nevertheless converge to neo open reading frame peptides (NOPs) from their translation products that result in common neoantigens in large groups of cancer patients. These disclosures are based, in part, on the identification of common, tumor-specific novel open reading frames resulting from frame-shift mutations. Accordingly, these disclosures provide novel tumor neoantigens and vaccines for the treatment of cancer. In some embodiments, multiple neoantigens corresponding to multiple NOPs can be combined, preferably within a single peptide or a nucleic acid molecule encoding one such single peptide. The advantage here is that a large percentage of patients can be treated with a single vaccine.
In detail, these disclosures demonstrate that within a single cancer type, the fraction of patients with a frame-shift in a subset of genes is much higher than the fractions identified when looking at all cancer patients. Inventors find that analysis of the data shows that frame-shift mutations in only a few genes are found in numerous patients:
- 5 genes together (GATA3, CDH1, MAP3K1, RUNX1, and TP53) are found in 20% of all breast cancers (WO2020022899).
- 4 genes together (BAP1, PBRM1, SETD2, and VHL) are found in 27% of all kidney cancers (WO2020022900).
- 8 genes together (APC, ARID1A, KMT2D, RNF43, SOX9, TCF7L2, TP53, and ZFP36L2) are found in up to 50% of all colorectal cancers (WO2020022902).
- 5 genes together (ARID1A, KMT2B, KMT2D, PIK3R1, and PTEN) are found in at least 30% of all uterine cancers (WO2020022901).
- In addition: TP53 frame-shift mutation peptides covering up to 4% of cancer patients, ARID1A frame-shift mutation peptides covering up to 3% of cancer patients, KMT2D frame-shift mutation peptides covering up to 2.14% of cancer patients, PTEN frame-shift mutation peptides covering up to 1.3% of cancer patients, KMT2B frame-shift mutation peptides covering up to 1.1% of cancer patients, CDKN2A frame-shift mutation peptides covering up to 0.6% of cancer patients (WO2020022903).
Methods for identifying neoantigens for the manufacture of mRNA vaccines
Moreover, in addition to patent families claiming cancer vaccines, patent family WO2020022897 provides methods for identifying neoantigens for preparing an off-the-shelf vaccine for treating cancer. Overall, this method involves the following steps:
- Identifying frame-shift mutations in the tumor DNA and/or RNA of a cohort of cancer patients in order to obtain a frame-shift library;
- Identifying at least one gene which is changed by a frame-shift mutation in the tumor DNA and/or RNA of one or more patients in the cohort of cancer patients to obtain a frame-shift gene;
- Identifying each novel open reading frame in both the +1 and -1 reading frame that overlaps with or is adjacent to the frame-shift location of the frame-shifted gene to obtain candidate novel open reading frame sequences;
- optionally when present, identifying each novel open reading frame in both the +1 and -1 reading frame that overlaps with or is adjacent to the frame-shift location for each alternative splicing construct of the frame-shift gene to obtain candidate novel alternative splicing open reading frame sequences;
- Combining each of the candidate open reading frame sequences and optionally the candidate novel alternative splicing open reading frame sequences of the frame-shift gene in a nucleic acid construct.
The figure below is a schematic overview of this method:
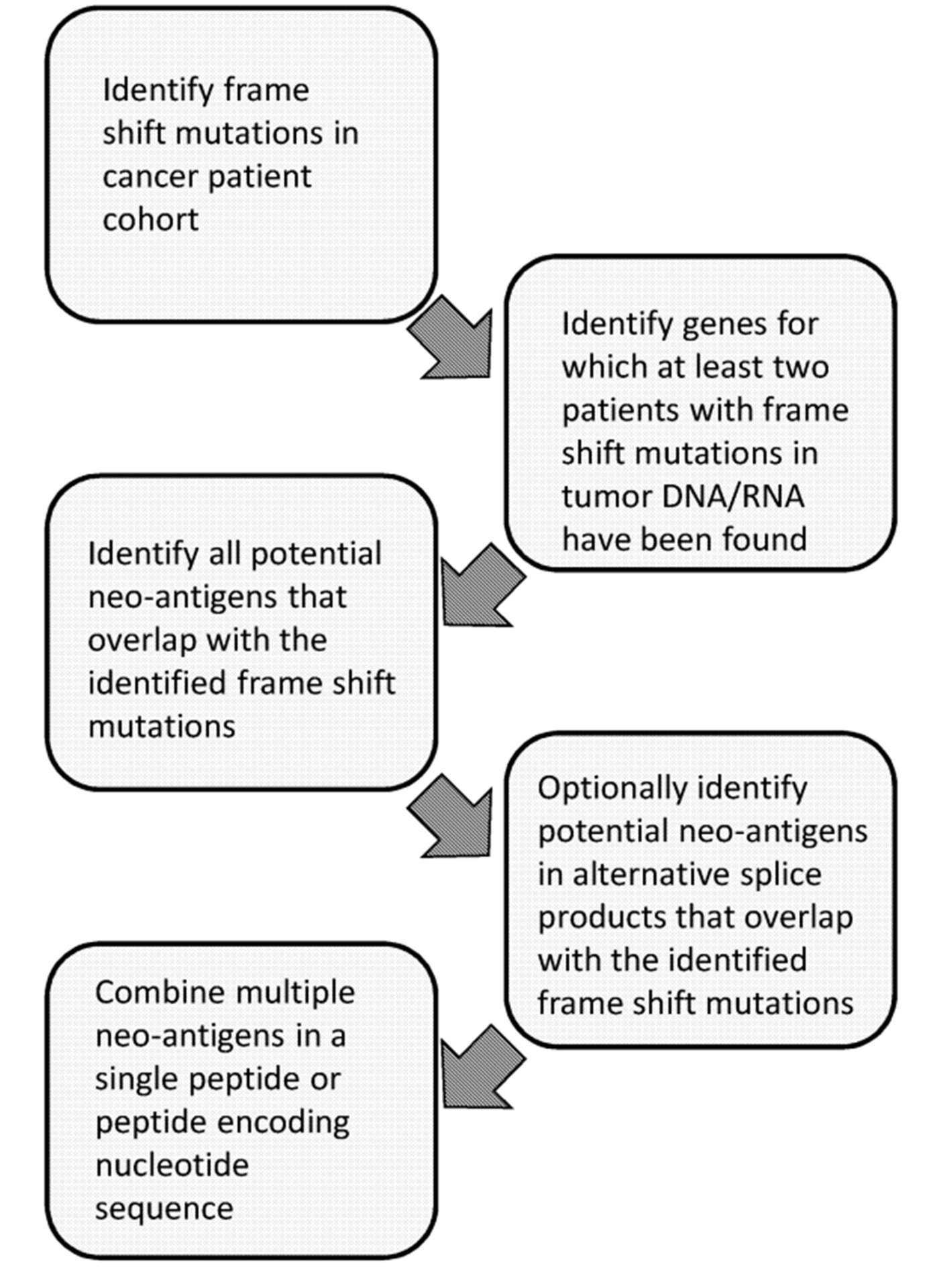
Figure 1: Schematic overview of methods for identifying neoantigens for preparing an off-the-shelf cancer vaccine (WO2020022897).
In addition to the method for identifying neoantigens, this patent family also claims a peptide comprising at least two amino acid sequences independently selected from the group consisting of SEQ ID Nos 1 to 4307. These amino acid sequences have been found through this method in tumors in cancer patients, and are the consequence of frame-shift mutations that have been introduced in the genome of the cancer cells of cancer patients.
Finally, the latest pending patent application (WO2021172990) filed by Frame Pharmaceuticals in 2020 relates to a new class of neoantigens, referred to as ‘hidden frames’, as well as methods for identifying such neoantigens. It is worth noting that this pending patent application relates to the FramePro platform developed by Frame Therapeutics. According to the company, Frame’s FramePro platform identifies structural changes within the cancer genome that give rise to new open reading frames. These new open reading frames result in novel proteins that are absent in healthy tissues and can thereby be recognized as foreign by the immune system.
This pending patent application gives interesting insights into the process behind the FramePro platform. As seen previously, inventors have screened over 10,000 sequenced tumors, of all major cancer types, to determine the fraction of tumors that have at least 1 frame-shift-caused encoded neopeptide of at least 10 amino acids (abbreviated as one ‘frame’), and also the fraction that has at least 2 frames, 3 frames, etc. Even when excluding microsatellite instable tumors (which contain many frame-shift indels), inventors find that the major fraction of tumors contains 1 or more frames. For example, in lung cancer, 95% of patients have at least 1 frame derived from frame-shift indels and 75% have a least 4 frames.
The method (named ‘FramePro’ or ‘reconstructed tumor genome mapping’) comprises the generation of a tumor-specific human reference genome, based on somatic and germline structural genome variations identified in a tumor sample, followed by mapping of long cDNA/RNA reads to the tumor-specific reference sequences. The method involves the following steps:
- Whole genome sequencing (WGS) of a tumor sample and a healthy sample from the individual as described further herein. Preferably, WGS of the tumor sample includes long-read sequencing. As demonstrated in the disclosure, long-read genome sequencing allows reconstruction of complex DNA rearrangements.
- Long-read RNA sequencing of RNA from at least one tumor sample. Preferably the RNA is selected or enriched for poly-(A) mRNA and/or 5’- CAP containing mRNA.
- Optionally performing short-read RNA sequencing on RNA from at least one tumor sample.
- Mapping the genomic sequences obtained to a human reference sequence to identify somatic structural genomic variations in the tumor sample. This step also distinguishes germline genetic variations (identified from the healthy tissues) from tumor-specific genetic variations (identified from the tumor tissue).
- Generating in silico a reconstructed tumor-specific reference genome comprising the identified somatic structural genomic variations. It is not necessary to generate a complete tumor-specific reference genome. Rather, contigs which span the structural genomic variations can be generated. Such contigs are generally around 100kb but can be longer, e.g., 300-400kb.
- Aligning the RNA sequences to the reconstructed tumor-specific reference genome. This step is useful when mapping RNA sequencing data to the genome.
- Determining the sequences of the full-length RNA transcripts encoded by the structural genomic variations. The disclosure provides that when the transcription/splicing machinery encounters a DNA rearrangement, it will often seek new splice sites resulting in an RNA transcript with a novel open reading frame. Based on the WGS and RNA sequencing data provided above, the sequence of these new RNA transcripts can be determined.
- Determining the predicted amino acid sequences encoded by the full-length transcripts of g). This method provides an improved pipeline for determining tumor neoantigens, in particular for neoantigens resulting from complex chromosomal rearrangements. This method can also be used to select for such tumor neoantigens by:
- Selecting, as candidate neoantigen sequences, sequences comprising about 10 contiguous amino acids of the predicted amino acid sequence of h), wherein at least four of the contiguous amino acids are not encoded in the germline genome of the individual.
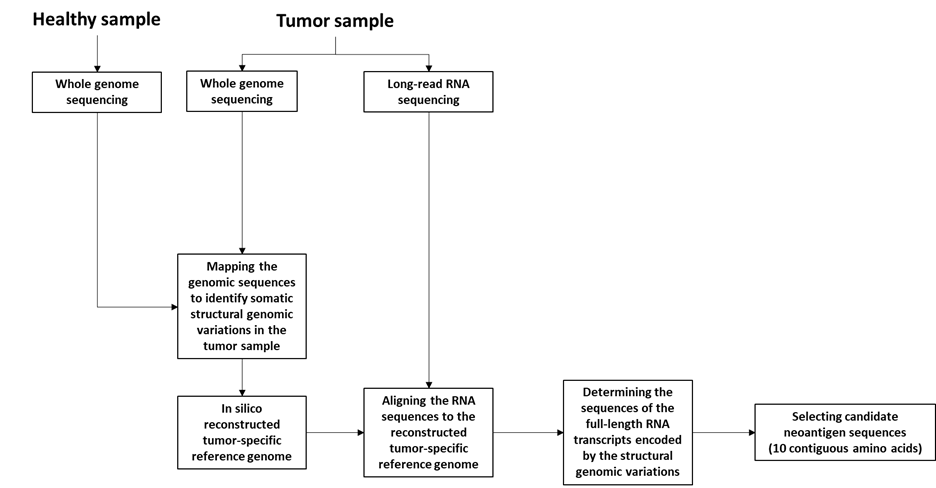
Figure 2: Schematic overview of FramePro method for identifying neoantigen sequences (adapted from WO2021172990).
Current legal status of CureVac’s recently acquired patent portfolio
From a legal point of view, there are currently no granted patents in its patent portfolio. Apart from its last patent family with one pending PCT patent application (WO2021172990) that is not yet extended to foreign countries, each patent family comprises pending patent applications in the US, Europe, India, Israel and Canada. Therefore, Asia is not covered by this patent portfolio.
According to the written opinion of the International Searching Authority, the closest prior art for pending patent applications claiming cancer vaccines are documents filed by Neon Therapeutics (WO2017173321) and by the Broad Institute & Dana Farber Cancer Institute (WO2016187508). Therefore, independent claims of the European patent applications were amended to specific identified amino acid sequences. It is likely that these pending patent applications will be granted as such.
Regarding the pending patent application providing methods for identifying neoantigens for preparing an off-the-shelf vaccine, it is mainly anticipated by a document filed by the Dana Farber Cancer Institute (WO2011143656). Despite the pending set of claims of the European patent application was amended, it is not certain that these modifications can restore the novelty and inventive step of the invention.
Concerning the last PCT pending patent application filed by Frame Cancer Therapeutics (WO2021172990), none of the prior art documents found disclose a method for identifying neoantigens comprising all the steps and features recited in claim 1. It is therefore likely the subject matter of the method for identifying neoantigens as described in the 1st claim can be granted without restrictive amendment relatively quickly, thus securing its FramePro platform in the US, Europe, Canada, Israel and India (the common designated countries for Frame Cancer Therapeutics).
In conclusion, all patent families filed by Frame Cancer Therapeutics are well detailed, giving several concrete examples, and are supported by a scientific article published in a peer-reviewed journal. Furthermore, several of its pending patent applications should be granted soon in the US and in Europe. The technology developed is therefore promising, particularly credible and shows solid assets for providing personalized therapeutic cancer vaccines that can be provided off the shelf. Therefore, the addition of Frame’s technology will aptly complement CureVac’s ability to identify and validate relevant neoantigens for its mRNA cancer vaccine programs.
KnowMade’s healthcare patent reports.
Press contact
contact@knowmade.fr
Le Drakkar, 2405 route des Dolines, 06560 Valbonne Sophia Antipolis, France
www.knowmade.com
About the author
Brice Sagot, CTO and co-founder of Knowmade, leads the Biotechnology and Life Sciences department. He holds a PhD in Molecular Biology from the University of Nice SophiaAntipolis (France).
About Knowmade
Knowmade is a Technology Intelligence and IP Strategy consulting company specialized in analysis of patents and scientific information. The company helps innovative companies and R&D organizations to understand their competitive landscape, follow technology trends, and find out opportunities and threats in terms of technology and patents.
Knowmade’s analysts combine their strong technology expertise and in-depth knowledge of patents with powerful analytics tools and methodologies to turn patents and scientific information into business-oriented report for decision makers working in R&D, Innovation Strategy, Intellectual Property, and Marketing. Our experts provide prior art search, patent landscape analysis, scientific literature analysis, patent valuation, IP due diligence and freedom-to-operate analysis. In parallel the company proposes litigation/licensing support, technology scouting and IP/technology watch service.
Knowmade has a solid expertise in Compound Semiconductors, Power Electronics, Batteries, RF Technologies & Wireless Communications, Solid-State Lighting & Display, Photonics, Memories, MEMS & Solid-State Sensors/Actuators, Semiconductor Manufacturing, Packaging & Assembly, Medical Devices, Medical Imaging, Microfluidics, Biotechnology, Pharmaceutics, and Agri-Food.
