SOPHIA ANTIPOLIS, France – July 07, 2021 | Natera was founded by Matthew Rabinowitz, Ph.D., and Jonathan Sheena, M. Eng, in 2004. This American company provides DNA testing across multiple clinical areas including women’s health, organ health, and oncology. This DNA testing, involving a noninvasive blood-based test, is carried out using its next-generation cfDNA technology platform. Natera offers proprietary genetic testing services to inform obstetricians, transplant physicians, oncologists and cancer researchers, including biopharmaceutical companies and genetic laboratories, through its cloud-based software platform. Its global footprint spans 90 countries worldwide.
Its technology platform combines novel molecular biology techniques with a suite of bioinformatics software that enables detection down to a single molecule in a tube of blood. With this platform, the company develops an accurate noninvasive prenatal test which is currently on the market (Panorama™), a tumor-specific assay for truly individualized cancer care (Signatera™) and best-in-class rejection assessment for kidney transplantation (Prospera™), among other transformative cfDNA tests. In March 2021, Natera announced that the US FDA had granted two Breakthrough Device Designations covering new intended uses of the Signatera™ molecular residual disease test in cancer.
Natera evaluates single-nucleotide polymorphisms (SNPs) – the 1% of DNA that makes us different from one another. The precision and accuracy of its technology is what enables Natera’s tests to distinguish between the DNA of a mother and a baby, or an organ transplant donor and a recipient.
In 2019, Natera introduced the Prospera™ test. Covered by Medicare (the US federal health insurance program), Prospera™ is a transplant rejection test that uses a blood draw to assess the risk of rejection of a transplanted kidney. It increases a provider’s ability to identify otherwise undetected rejection that might lead to kidney loss. Catching transplant rejection as soon as possible can help providers develop a treatment plan to best protect the donated kidney.
The Prospera™ test leverages Natera’s core single-nucleotide (SNP)-based massively multiplexed PCR (mmPCR) technology to identify allograft rejection noninvasively and with high precision and accuracy, without the need for prior donor or recipient genotyping. The test works by measuring the fraction of donor-derived, cell-free DNA (dd-cfDNA) in the recipient’s blood. Upon cell injury, more dd-cfDNA is released from the kidney. With a 95% negative predictive value, Prospera™ misses ~3x fewer rejections than serum creatinine, a classic test for kidney transplant monitoring. It may be used by physicians considering the diagnosis of active rejection, helping to rule in or out this condition when evaluating the need for diagnostic testing or the results of an invasive biopsy. Prospera™ has been clinically and analytically validated for performance regardless of donor relatedness, rejection type and clinical presentation.
The test performance characteristics have been determined by the CLIA (Clinical Laboratory Improvement Amendments)-certified laboratory performing the test. The test has not been cleared or approved by the US Food and Drug Administration (FDA).
• September 2020: Natera announced the publication of a prospective, randomized controlled trial demonstrating the clinical utility of the Prospera™ test. The study concluded that practicing nephrologists who used the Prospera™ test detected more cases of rejection and made better clinical decisions than physicians in the control group. The study evaluated kidney transplant patients in typical scenarios seen by nephrologists in routine practice (“Randomized clinical trial of a novel donor-derived cfDNA test to detect rejection in CPV-simulated renal transplant patients”, International Urology and Nephrology, 2020).
• April 2021: Natera received pathway to future coverage for the Prospera™ donor-derived, cell-free DNA test to determine transplant rejection status in multiple organs. The Centers for Medicare & Medicaid Services (CMS) Molecular Diagnostics Program (MoLDX) published a future effective local coverage determination (LCD) that identifies and establishes this pathway to coverage in a broad range of solid organ transplants. This new LCD adds to existing CMS coverage for the Prospera™ test in kidney transplant rejection assessment.
Since its creation in 2004, Natera has filed 64 patent families, of which 36 in the field of circulating DNA (Circulating DNA/RNA – Patent landscape 2021), including 259 patent documents published between 2011 and 2020. In the circulating DNA field, 79% of Natera’s patents are currently alive, and among them, 60% are pending applications. The company’s portfolio is mainly focused on prenatal and cancer diagnostics, and is 5 years old on average. Natera has a worldwide IP strategy, with granted patents and pending applications in the US, Europe and Asia (mainly Japan and China).
Among these 36 patent families, Natera owns 1 patent family focused on transplantation (Methods for detection of donor-derived cell-free DNA – WO2020/010255), and 3 others, which are more general, where circulating DNA can be used for prenatal diagnosis, cancer and transplantation (Methods for simultaneous amplification of target loci – US10597723; Methods and systems for calling ploidy states using a neural network – WO2020/018522; Improved liquid biopsy using size selection – WO2020/214547).
The patent family focused on transplantation (WO2020/010255) is closely related to the Prospera™ test features. It provides methods for determining the status of an allograft within a transplant recipient from genotypic data measured from a mixed sample of DNA (DNA from both the transplant recipient and the donor). The mixed sample of DNA may be enriched at a plurality of polymorphic loci in a way that minimizes the allelic bias, for example using massively multiplexed targeted PCR. This patent family, filed in July 2019, comprises a pending PCT application (WO2020/010255) with pending applications in Europe (EP3818177), China (CN112752852) and Brazil (BR112020027023). These pending applications are recent, and could be granted in the future. However, the written opinion of the International Searching Authority (ISA) discloses that the claims could be modified because the first claim lacks novelty, as do a quarter of the claims, and not all are inventive.
The first claim of the PCT application (WO2020/010255) is a method of quantifying the amount of donor-derived, cell-free DNA (dd-cfDNA) in a blood sample from a transplant recipient, which involves:
a) extracting DNA from the blood sample of the transplant recipient, wherein the DNA comprises donor-derived, cell-free DNA and recipient-derived, cell-free DNA;
b) performing targeted amplification at 500-50,000 target loci in a single reaction volume using 500-50,000 primer pairs, wherein the target loci include polymorphic loci and non-polymorphic loci, and wherein each primer pair is designed to amplify a target sequence of no more than 100 bp; and
c) quantifying the amount of donor-derived, cell-free DNA in the amplification products.
According to the ISA, regarding novelty, a patent family filed by Natera in 2015 (US10316362) already provides methods combining multiplex PCR and sequencing, such as high throughput sequencing. It discloses methods that permit the targeted amplification of over a hundred to tens of thousands of target sequences (e.g. SNP loci) from a nucleic acid sample, such as genomic DNA obtained from plasma. In short, it is likely that Natera will restrict its claims for national patent applications (WO2020/010255) to kidney transplants, for example.
Regarding the technology described, this patent family relates to methods for determining the status of an allograft from a mixed sample of DNA. More precisely, circulating free DNA from plasma was extracted using a cfDNA kit (Qiagen). The amount of cfDNA was quantified and a library preparation was accomplished (Natera Panorama Library Prep Kit). The amplified library was then purified, quantified again and a quality control step was performed. The samples were then pooled for sequencing, purification, quantification and quality control. The percentage of donor-derived, cell-free DNA in the transplant recipient plasma was determined using massively multiplexed PCR, which targeted 13,392 single-nucleotide polymorphisms (SNPs), followed by NGS sequencing on a HiSeq2500 machine (Illumina), without the need for donor genotypes. An increase in the level of dd-cfDNA was correlated with rejection and transplant injury status. An amount greater than 1% of dd-cfDNA indicates that the transplant is undergoing acute rejection (antibody or T cell mediated transplant rejection).
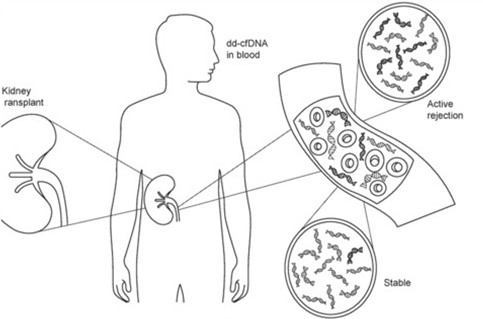
Figure 1 shows how DNA released from transplanted kidneys into the bloodstream is elevated in acute graft rejection (WO2020/010255).
Patent family WO2020/010255 describes several advantages over existing technologies:
Firstly, dd-cfDNA is an effective prognostic marker. Current standard-of-care clinical options to monitor kidney health in transplant recipients include protocol-biopsies and assessing dynamic changes in serum creatinine and other parameters such as proteinuria and levels of immunosuppressive drugs. Although protocol-biopsies are considered the “gold standard”, their clinical utility is significantly limited, due to invasiveness, cost, inadequate sampling and poor reproducibility. Serum creatinine, the current standard-of-care marker for screening renal allograft dysfunction and indicating when biopsy and histological evaluation of renal tissue is warranted, is a poor marker, due to its low sensitivity and specificity. Moreover, creatinine is a lagging indicator of renal injury; by the time serum creatinine levels increase, the allograft has already undergone severe and irreversible damage. In one example (WO2020/010255), it was observed that median dd-cfDNA was significantly higher in the acute rejection (AR) group versus the non-AR group. In contrast to dd-cfDNA, evaluation of creatinine levels did not appear to have as much discriminatory ability for differentiating between AR and non-AR groups (Fig. 2A-B).
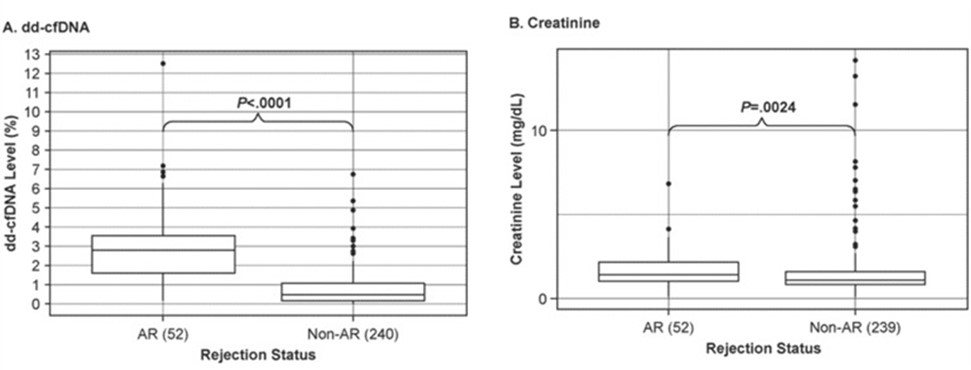
Figure 2: Discrimination of active rejection by dd-cfDNA (A) versus creatinine (B). Boxes indicate interquartile range (25th to 75th percentile); horizontal lines in boxes represent medians; dots indicate outliers >1.5 times the upper quartile value. P- values for dd-cfDNA adjusted using a Kruskal-Wallis rank sum test followed by Dunn multiple comparison tests with Holm correction; P-values for creatinine adjusted via Tukey’s test (WO2020/010255).
Furthermore, several methods have been improved compared to the state of the art. The first technical improvement made in this patent family is a size selection step to eliminate background noise. After blood draw and before DNA extraction, blood cells within a blood sample may burst and shed long fragments of DNA into the sample, which would increase the total amount of cfDNA and background noise. To reduce such background noise, and based on the observation that dd-cfDNA is typically shorter than DNA shed from a transplant recipient blood cell, a size selection step is applied to select shorter cfDNA. Moreover, other technical improvements are that the assay can filter PCR and NGS errors through advanced error modeling, and uses a very high number of SNPS (>13,000) with advanced SNP selections. The method uses highly efficient, highly multiplexed targeted PCR to amplify DNA followed by high throughput sequencing to determine the allele frequencies at each target locus. This approach allows multiplexing of at least 10,000 primers in a single pool, with the resulting amplified DNA comprising a majority of DNA molecules that, when sequenced, will map to targeted loci. In one example (WO2020/010255), analysis of performance estimates demonstrated that the massively multiplexed-PCR-NGS method was able to discriminate active from non-active rejection status with high sensitivity (92.3%) and specificity (72.9%) at the acute rejection cutoff of >1% dd-cfDNA. Serum creatinine levels were less discriminatory, with 42.3% sensitivity and 83.7% specificity.
Finally, one of the important points of the developed method is that there is no need for donor genotypes. One barrier to widespread clinical use of dd-cfDNA as a diagnostic tool for monitoring organ transplant has been the limitations in measuring dd-cfDNA in certain cases such as when the donor genotype is unknown or when the donor is a close relative. Given the design of the assay used here, it is possible to quantify dd-cfDNA without prior recipient or donor genotyping. Further, there is no need for a computational adjustment based on whether the donor is related to the recipient. In one example (WO2020/010255), evaluation of dd-cfDNA levels by donor type revealed that regardless of donor type (living related, living non-related, deceased non-related), dd-cfDNA levels were similar across all donor types within the AR and non-AR categories.
According to Natera, patent family WO2020/010255validates the use of dd-cfDNA in the blood as an accurate marker of kidney injury/rejection. This rapid, accurate, and noninvasive technology may offer detection of significant renal injury in select patients better than the current standard of care and therefore offer the potential for better management and survival of kidney allografts and recipient renal function.
However, other tests to prevent kidney transplant rejection are also present on the market. First, Exosome Diagnostics develops ExoTRU (Exosome Transplant Rejection Urine): a noninvasive, multigene, urine-based exosomal mRNA assay. This urine test, based on exosome, has very different characteristics from Natera’s. Next, NephroSant provides QSant™, another urine-based test. Statistical models based upon the measurements of the six biomarkers (cell-free DNA, methylated-cell-free DNA, clusterin, CXCL10, creatinine and total protein) generated a renal transplant Q-Score that reliably differentiated stable allografts from acute rejections in patients. Although this test is closer to Natera’s, it is still quite different (urine, 6 biomarkers, algorithm).
Finally, CareDx, an American company specialized in transplantation care, supplies AlloSure®, a dd-cfDNA test for noninvasive transplant surveillance. It measures cfDNA and uses SNPs to distinguish between donor and recipient. It can quantify increasing levels of dd-cfDNA, serving as a leading indicator of graft injury. The CareDx and Natera tests to assess the risk of rejection of a transplanted kidney are very similar – which could explain the rivalry between these two companies. In 2019, a straw man company (which Natera is probably behind) filed an opposition against a CareDx patent family (EP3117012), and CareDx along with Stanford University sued Natera for patent infringement that same year. They claimed that Natera had infringed US Patent US9845497by developing and commercializing the Prospera™ test. In 2020, Natera sued CareDx for patent infringement (AlloSure®). Moreover, in 2020, Natera filed suit against CareDx for false advertising, alleging CareDx used false and misleading claims to deceive physicians about the performance capabilities of its AlloSure test, in violation of various laws prohibiting unfair competition and deceptive trade practices. Although all the proceedings are still pending, the two tests and the various patent families seem very close. In view of the commercial stakes, there appears to be a long battle coming between Natera and CareDx.
To conclude, Natera is developing an innovative technology with its Prospera™ test which is very important in the field of transplantation. Indeed, this test is simpler and less invasive than tissue biopsy and can measure more relevant biomarkers (i.e. dd-cfDNA versus creatinine). However, the medico-economic stakes are high. Natera is not the only company in this area of activity and the IP battle with CareDx is tough. This year will surely be decisive for both companies in the transplantation care field.
Take advantage of KnowMade’s life sciences patent landscape reports.
Press contact
contact@knowmade.fr
Le Drakkar, 2405 route des Dolines, 06560 Valbonne Sophia Antipolis, France
www.knowmade.com
About our analysts
Fabienne works at Knowmade in the field of Biotechnology and Life Sciences. She holds a PhD in Molecular and Cellular Biology from the IPMC (Nice, France). She also holds a Master of Business Management from IAE (Nice, France) and she previously worked in the pharmaceutical industry.
About Knowmade
Knowmade is a Technology Intelligence and IP Strategy consulting company specialized in analysis of patents and scientific information. The company helps innovative companies and R&D organizations to understand their competitive landscape, follow technology trends, and find out opportunities and threats in terms of technology and patents.
Knowmade’s analysts combine their strong technology expertise and in-depth knowledge of patents with powerful analytics tools and methodologies to turn patents and scientific information into business-oriented report for decision makers working in R&D, Innovation Strategy, Intellectual Property, and Marketing. Our experts provide prior art search, patent landscape analysis, scientific literature analysis, patent valuation, IP due diligence and freedom-to-operate analysis. In parallel the company proposes litigation/licensing support, technology scouting and IP/technology watch service.
Knowmade has a solid expertise in Compound Semiconductors, Power Electronics, Batteries, RF Technologies & Wireless Communications, Solid-State Lighting & Display, Photonics, Memories, MEMS & Solid-State Sensors/Actuators, Semiconductor Manufacturing, Packaging & Assembly, Medical Devices, Medical Imaging, Microfluidics, Biotechnology, Pharmaceutics, and Agri-Food.
